Use examples
Running on Command-line
Merging reconstructions built using different templates
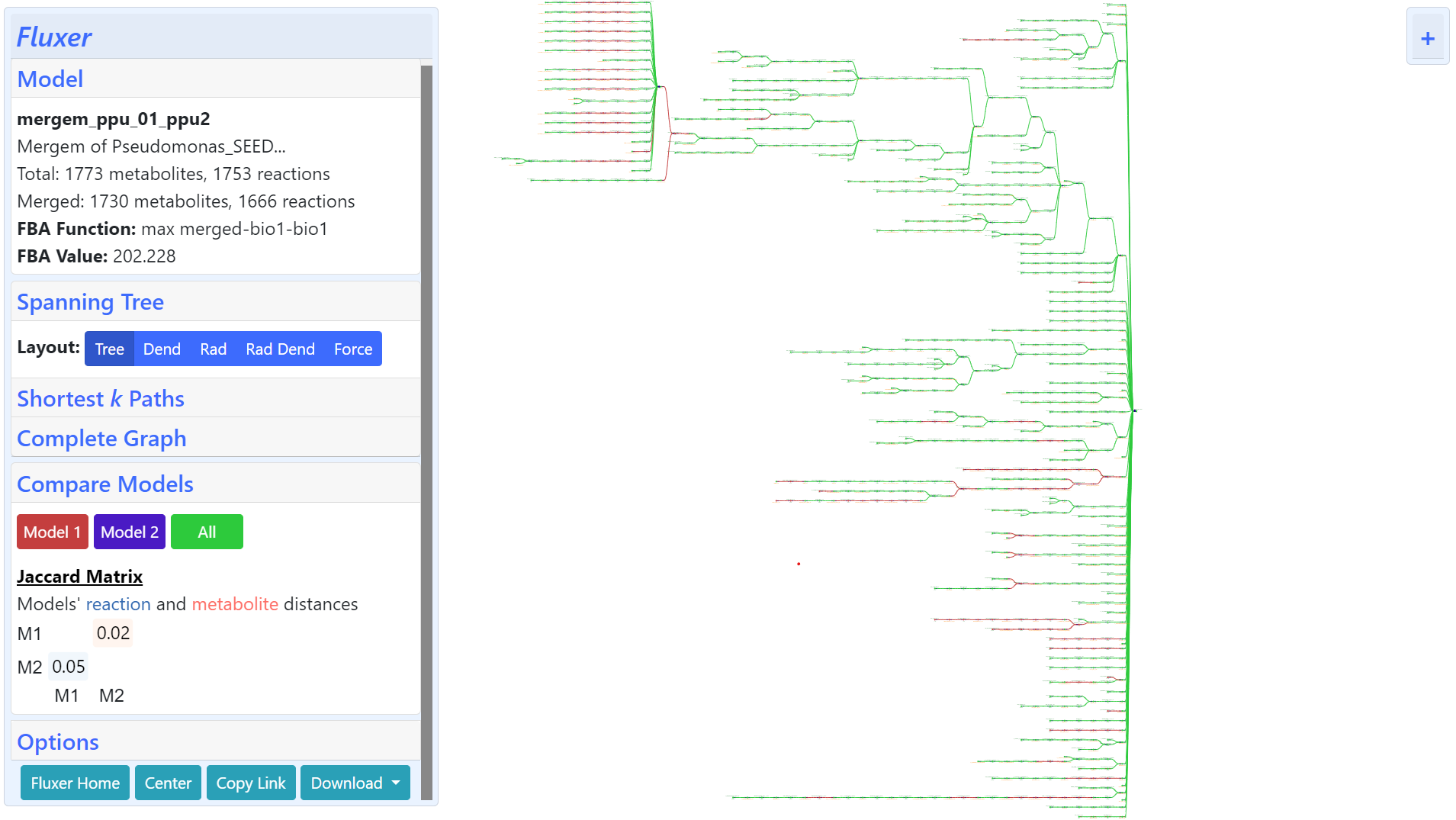
To compare the effect of reconstruction parameters, we merged two P. putida draft reconstructions built on ModelSEED using either a gram-positive or core template.
mergem -v MS1_PPU.sbml MS2_PPU.sml -o MS1_MS2_merged_PPU.xml
Results from merging the two reconstructions on the command-line using mergem are shown below.
Merging models complete. Merged model saved as MS1_MS2_merged_PPU.xml
Jaccard distance matrix: [[0, 0.02425267907501405], [0.049657534246575374, 0]]
Metabolites merged: 1730
Reactions merged: 1666
The figure below shows the same merging on the web-application Fluxer and can be accessed here.
Importing Python package
Studying model versions
Different versions of models can be compared to analyze elements that were added or removed during update. The results of comparing three versions of a P. putida KT2400 model using mergem on the Python console are shown below:
import mergem
input_models_list = ['models/iJN1463.xml', 'models/iJN746.xml', 'models/MNX_iJN746.sbml']
merge_results = mergem.merge(input_models_list, set_objective='merge')
print('Number of metabolites in merged model: ', len(merge_results['merged_model'].metabolites))
print('Number of reactions in merged model: ', len(merge_results['merged_model'].reactions))
print("Jaccard matrix: \n", merge_results['jacc_matrix'])
print('Number of metabolites merged between input models: ', merge_results['num_met_merged'])
print('Number of reactions merged between input models: ', merge_results['num_reac_merged'])
Running the above script produces the following output:
Number of metabolites in merged model: 2186
Number of reactions in merged model: 3001
Jaccard matrix:
[[0, 0.5976168652612283, 0.5986270022883295], [0.6765588529509836, 0, 0.00658616904500553], [0.679, 0.01526717557251911, 0]]
Number of metabolites merged between input models: 1783
Number of reactions merged between input models: 2024
The same result including the Jaccard matrix can be visualized on Fluxer as shown below:
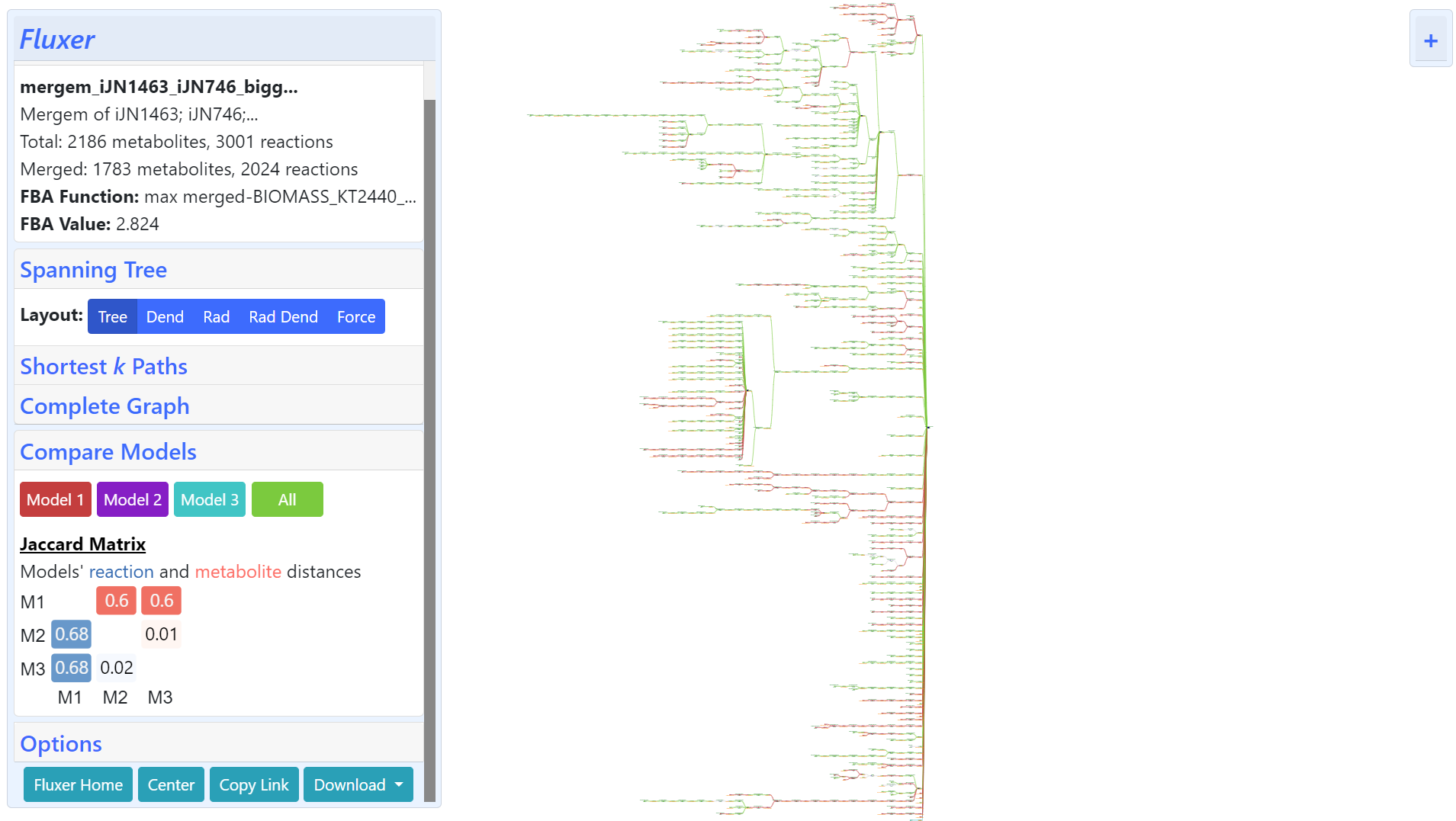
The above Fluxer result can be accessed here.